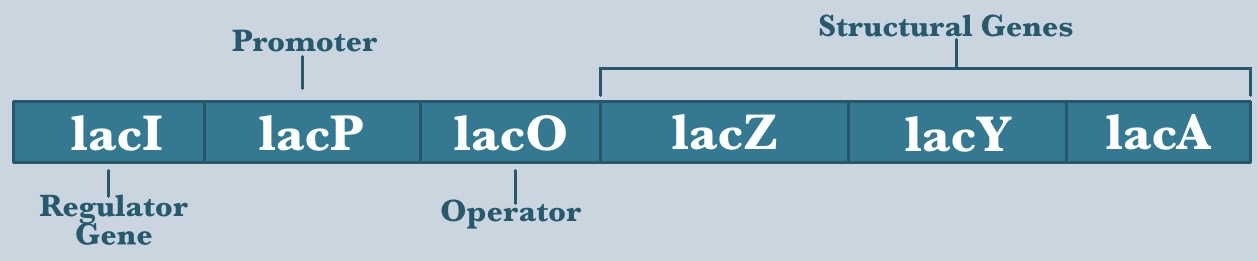
LacOp: An online simulation on Prokaryotic Transcriptional Regulation
This simulation approximates the function of the E coli “Lactose Operon” in a single E. coli. Use the tabs below to configure an initial sugar concentration, select desired mutations and then click the button to “Run Pathway & Make Graph”. For example, (e.g. select Sugars, and set Lactose outside to 100 units, with the other sugars set to ‘0’), and select “Run…’. Each run will create a dynamic pathway simulation with these inputs on our server and graph which the concentrations of key molecules through time. By rerunning the pathway and modifying sugar levels and/or mutations, students will gain a deeper understanding of how the lac operon is transcriptionally regulated.
For initial simplicity, this simulation runs with a defaults that simulates the wild-type Lac Operon, with the exception that “CAP” has been initially disabled. When students are ready for additional complexity, they can activate CAP using the “CAP-cAMP complex” tab. Students can also test partial diploid strains using the “Plasmid” tab.
The Lac Operon is most famous for its role in helping geneticists understand transcriptional gene regulation in prokaryotes. In this exercise, you will use an interactive online simulation called ‘LacOp’ to model the metabolism of a single E.coli cell. Our goal is to help you understand the dynamics of transcriptional gene regulation in a more fun and effective way than you could from a textbook.